Data splitting challenge
Mowing detection
One of the examples where we meet the splitting challenge is the mowing detection task. The goal of mowing detection is to predict the time at which the grass on the field was cut. Thus, as part of our mowing detection project, each year we collect field books from farmers and field reports from the inspectors of the local paying agency. The received data is converted and reviewed manually, and some of the ground truth is produced from the manual labelling.
The labels would differ in trustworthiness, depending on the source (farmer field books, inspector field reports, any of the former with manually adjusted dates, or fully manual labelling). Since inspector field reports are the most reliable source, we would use most of them for the validation and test set. However, we would need at least some of them to be present in the training dataset as well. Additionally, each dataset is expected to have as well balanced classes distribution as possible, perhaps with additional filtering to randomly drop least trustworthy samples from the over-represented class.
Considering the aforementioned conditions, let’s say we would like to have 70% of the labelled data for training, 20% for validation and 10% for testing. For validation and testing, we would only use instances from inspector field reports and farmer field books with tweaked dates. For training, we would use data from all sources, including the ones from inspector field reports and tweaked field books which were left over from the test and validation datasets.
Crop classification
Another task we are dealing with is crop classification. We would like to detect the crop type of agricultural fields out of 28 possible classes. Similarly to the mowing detection we have different sources for labels, some of which have been provided by the local Agricultural Registers and Information Board, some from drone observations. For crop classification, class balance distribution plays the core role. In order to mitigate the issue of an unbalanced dataset, undersampling and oversampling can be used. Undersampling and oversampling should be available for the training subset, while for testing we would use some of the fields with labels of high confidence. Some of the classes might have a poor representation, due to which general split ratios might leave the validation or test dataset without any samples, whereas we need to ensure that all datasets have enough samples.
Thus, the requirements for splitting are the following. We would like to have 70% / 20% / 10% splits, ensuring that for smaller representing classes at least one instance is present in all sets. Additionally for the test set we would like to have the list of high confidence instances together with random leftover samples that added up to 10% of the whole data.
Generic and configurable
While such processing chains can be implemented, we have found it tricky to have it generic and configurable enough to cater for all sorts of projects with different (and sometimes rapidly changing) requirements.
Current solution
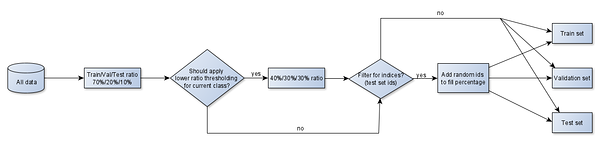
Currently we have separate implementations for mowing detection and crop classification, both of which take input parameters from a config file. The config file is basically python code and supports the definition of custom filter functions for datasets. For each dataset, the current solution invokes custom filters (if any) and then performs random sampling of data indices, leaving the rest of the samples for the next datasets. The samples which have been filtered out, are also left for the next datasets, for each dataset might have a different filter.
The reason why we prefer to use data / sample indices instead of data directly, is to have a layer of abstraction. This way the splitting logic could be agnostic of data type. It would not matter whether a single sample / instance is a raster image, an image time-series or time-series of parameter values which have been averaged over a pre-defined geometry.
For multiclass applications such as crop classification, data indices are sampled separately for each class within each dataset. The splitting also supports capping of samples for classes which are represented too well. However, if there are too few samples per class, a low threshold can be applied such that a different split ratio would be used. For instance, in the case of 70% training, 20% validation and 10% testing dataset with just 9 samples in one of the classes, we might end up with 7 samples in the training dataset, 2 samples in validation and 0 in the test set. To mitigate the issue, we could have the ratios adjusted to 40% training, 30% validation and 30% testing for classes with less than 100 samples.
Ideas for future developments
Instead of project-specific implementation of the splitting logic, we would prefer to have a generic framework for graph-based data splitting with support for cross-validation and bagging. Please let us know if there is such a framework already out there, or if there would be community interest in developing the framework.